2. Complete Example¶
Here is the source code of the examples/ex2_complete.py you ran in Getting Started.
examples/ex2.py
import argparse
import subprocess
import sys
from cosmos.api import (
Cosmos,
Dependency,
draw_stage_graph,
draw_task_graph,
pygraphviz_available,
py_call,
)
def echo(word, out_txt):
with open(out_txt, "w") as fp:
fp.write(word)
def cat(in_txts, out_txt):
subprocess.run(f"cat {' '.join(map(str, in_txts))} > {out_txt}", shell=True, check=True)
def word_count(in_txts, out_txt, chars=False):
c = " -c" if chars else ""
input = " ".join(map(str, in_txts))
subprocess.run(f"wc{c} {input} > {out_txt}", shell=True, check=True)
def recipe(workflow):
# Create two Tasks that echo "hello" and "world" respectively (source nodes of the dag).
echo_tasks = [
workflow.add_task(func=echo, params=dict(word=word, out_txt=f"{word}.txt"), uid=word, mem_req=10,)
for word in ["hello", "world"]
]
# Split each echo into two dependent Tasks (a one2many relationship).
word_count_tasks = []
for echo_task in echo_tasks:
word = echo_task.params["word"]
for n in [1, 2]:
cat_task = workflow.add_task(
func=cat,
params=dict(in_txts=[echo_task.params["out_txt"]], out_txt=f"{word}/{n}/cat.txt",),
parents=[echo_task],
mem_req=10,
uid=f"{word}_{n}",
)
# Count the words in the previous stage. An example of a simple one2one relationship.
# For each task in StageA, there is a single dependent task in StageB.
word_count_task = workflow.add_task(
func=word_count,
# Dependency instances allow you to specify an input and parent simultaneously.
params=dict(
in_txts=[Dependency(cat_task, "out_txt")], out_txt=f"{word}/{n}/wc.txt", chars=True,
),
mem_req=10,
# parents=[cat_task], <-- not necessary!
uid=f"{word}_{n}",
)
word_count_tasks.append(word_count_task)
# Cat the contents of all word_counts into one file. Only one node is being created who's
# parents are all of the WordCounts (a many2one relationship, aka a reduce operation).
summarize_task = workflow.add_task(
func=cat,
params=dict(in_txts=[Dependency(t, "out_txt") for t in word_count_tasks], out_txt="summary.txt",),
parents=word_count_tasks,
stage_name="Summary_Analysis",
mem_req=10,
uid="",
) # It's the only Task in this Stage, so doesn't need a specific uid
def main():
p = argparse.ArgumentParser()
p.add_argument("-drm", default="local", help="", choices=("local", "awsbatch", "slurm", "drmaa:ge", "ge"))
p.add_argument("-q", "--queue", help="Submit to this queue if the DRM supports it")
args = p.parse_args()
cosmos = Cosmos("cosmos.sqlite", default_drm=args.drm, default_max_attempts=2, default_queue=args.queue)
cosmos.initdb()
workflow = cosmos.start("Example2", skip_confirm=True)
recipe(workflow)
# any parameters that start with out_ are output directories, and will be created if
# the user calls workflow.make_output_dirs
workflow.make_output_dirs()
workflow.run(max_cores=10, cmd_wrapper=py_call)
# Noting here that if you wanted to look at the outputs of any Tasks to decide how to generate the rest of a DAG
# you can do so here, proceed to add more tasks via workflow.add_task(), and then call workflow.run() again.
# Yes, it does require running all Tasks in the dag to get the outputs of any Task, and we hope to address
# that limitation at some point in the future.
if pygraphviz_available:
# These images can also be seen on the fly in the web-interface
draw_stage_graph(workflow.stage_graph(), "/tmp/ex1_task_graph.png", format="png")
draw_task_graph(workflow.task_graph(), "/tmp/ex1_stage_graph.png", format="png")
else:
print("Pygraphviz is not available :(")
sys.exit(0 if workflow.successful else 1)
if __name__ == "__main__":
main()
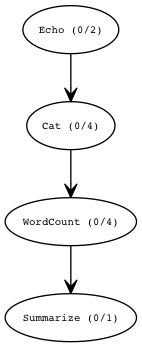
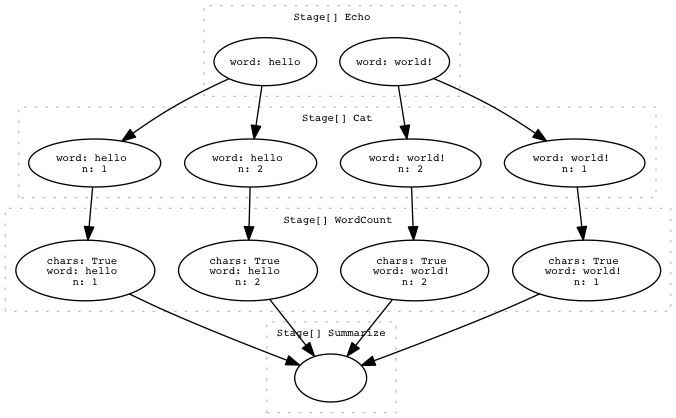
Here’s the DAG that was created. If you use the web-interface, the nodes are convenient links you can click to jump straight to the debug information that pertains to that Task or Stage.
The Stage Graph is a high level overview. Often the DAG of Tasks is too large for a visualization to be useful: